Author information
Annette Damert*
Download
Department of Primate Genetics, German Primate Center,
Leibniz Institute for Primate Research, Göttingen, Germany
Leibniz Institute for Primate Research, Göttingen, Germany
Abstract
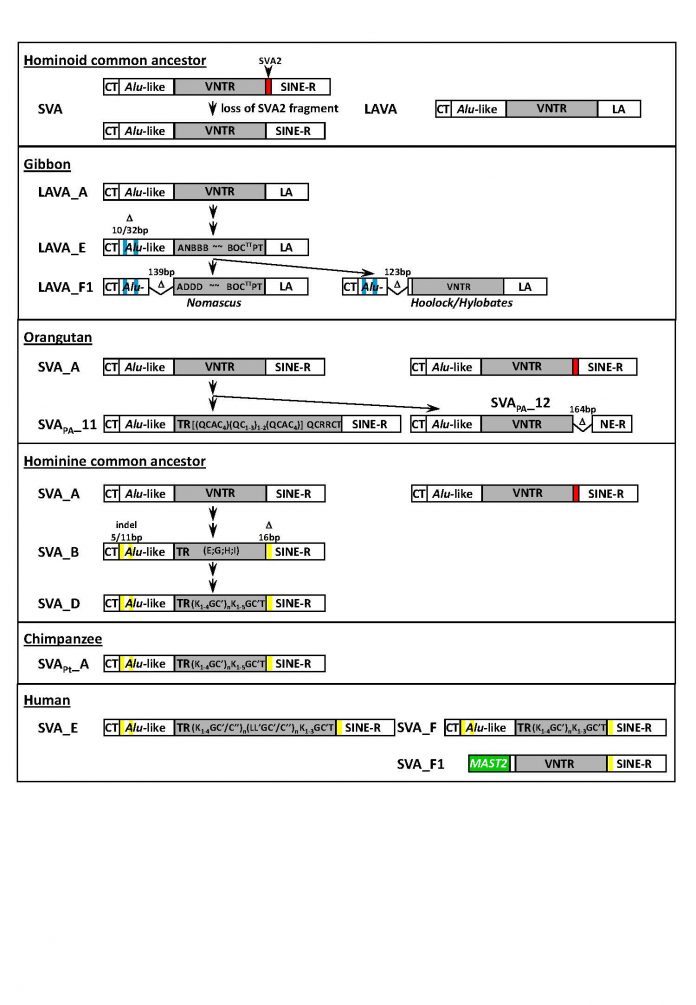
SVA (SINE-R-VNTR-Alu) and LAVA (L1-Alu-VNTR-Alu) are composite non-autonomous non-LTR retrotransposons specific to hominoid primates. They are mobilized by the LINE-1-encoded protein machinery in trans. SVA insertions can cause disease in humans. Both SVA and LAVA contribute to genome evolution by co-mobilization of non-retrotransposon sequences at their 5’ and 3’ ends.
The origin of SVA and LAVA can be traced back to a common ancestor present in the genomes of old world monkeys – SVA2. The shared 5’ Alu-like domain and the variant 3’ ends have been acquired through splicing. The evolutionary most recent addition to the family of VNTR (Variable Number of Tandem Repeats) – containing retrotransposons are human SVAs that have co-opted an exon of a host gene as their 5’ end.
Here I review current knowledge about the assembly of SVA/LAVA and their evolution in hominoid primates. Lineage- and family-specific structural differences will be discussed in the context of mobilization and host repression of expression.
(Received 14 March, 2018)
Keywords
retrotransposon, SVA, LAVA, evolution