Author information
Andrei Shutov1*, Irina Kakhovskaya1, Karl Wilson2
Download
1State University of Moldova, MD-2009 Chişinău, Moldova
2State University of New York at Binghamton, NY 13902-6000 Binghamton, NY, USA
2State University of New York at Binghamton, NY 13902-6000 Binghamton, NY, USA
Abstract
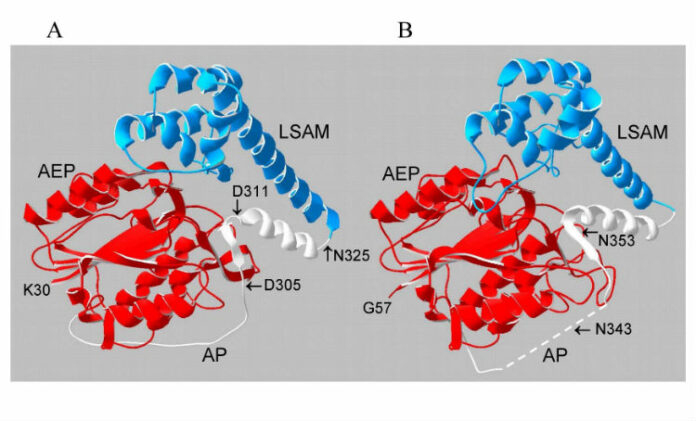
Evolutionary analysis of eukaryote legumains was conducted using sequences of their precursors composed of Asparaginyl EndoPeptidase (AEP) domain, Legumain Stabilization and Activity Modulation (LSAM) domain, and Activation Peptide (AP) in between. Three basic clusters were observed: ancient (from Parabasalia to Stramenopiles), plant and animal legumains. Early described mechanisms of auto-maturation of plant and animal legumains were compared in relation to their crystal structures and models. In both cases the auto-maturation starts from cleavage of Asn/Asp-flanked peptide bonds at central (plants) or C-terminal (animals) parts of the AP peptide resulting in detachment of the LSAM domain. It is finalized by cleavages of additional Asn/Asp-flanked bonds inside the AP peptide region that proximately block the active site Asn, His and Cys residues. In this way the active site triad becomes released, and complete activation of plant legumains occurs. Possibly, these additional cleavages might be essential for activation of animal legumains as well.
(Received 26 March 2019; accepted 5 November 2019)
Keywords
legumain, evolution, gene exon/intron structure, tertiary structure, activation